博文
could it flip?
|||
The GPCRDOCK 2013 contest hosted by Scripps Institute was finished months ago with the released of 5HT1B and 5HT2B crystal structures bound with agonist molecule Ergotamine. I also attended this event and compared my models with the crystal structure.
My models were built in Modeller templated with agonist bound beta1 adrenergic receptor crystal(PDB: 4AMJ). The sequence alignment was finished in clusterW2 and conserved residues and motifs were manually refined. The final alignment was end up with 40% sequence identity and 61% sequence similarity, both of which are pretty high.
Superimposing refined homology model and corresponding crystal structure indicated that the RMSD for both TMs backbone and sitechain in binding pocket were below 1.5 Å.

In principle, we should obtain very promising binding mode based on such good quality model, especially considering the target ligand is not so flexible. However, docking results from Glide showed that almost all top scored poses were far from the position observed in crystal structure (with RMSD>3.0Å).
What's happening?
Detailed analysis showed that the ligand structure Ergotamine downloaded from PUBCHEM (CID 8223) is deportonated. Ligand preparation steps retained the same absoulute configuration of each atoms. It is noticed that a N-methyl group adopted S configuration in crystal structure, while it is R configuration in the Ligpep output!
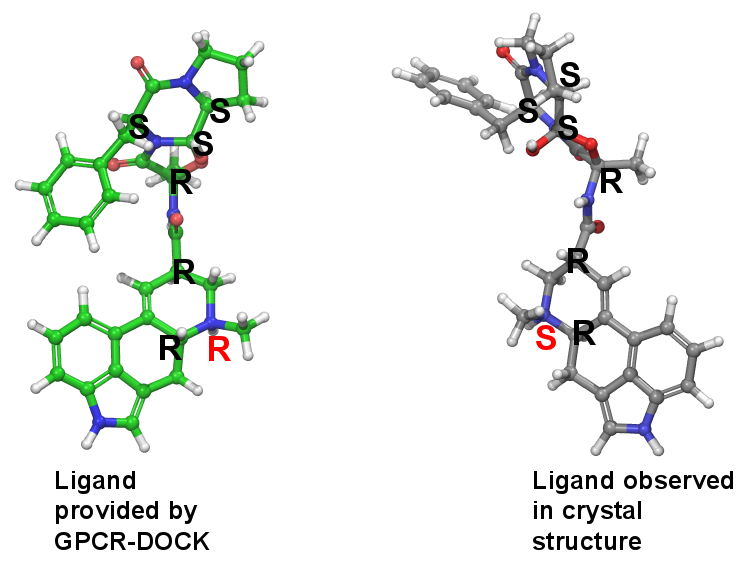
Obviously the problem comes from the Ligprep step. It is known to all, generally it is not so easy for protonated N-methyl group flip. Now the next question is: could the N-methyl group trun over at room temperature when it is deprotonated? If yes, the configuration can be changed when Ergotamine binds with the receptor. Otherwise, we are afraid that someone made mistakes in providing the accurate contest informations.
We may not answer this question simply by eye, although there is so called "nitrogen invention" in chemistry. QM calculations can help us finding the answers:
Inverted geometry was obtained at N and the planar configuration at
PBE0-dDsC/def2-TZVP//B3LYP-dDsC/6-31G*
The inverted configuration at N is computed to be 1.8 kcal/mol lower than the original one, and the barrier from the original to the constrained planar is 5.6 kcal/mol.
first of all, the electronic energy difference does not strictly correspond to a free energy difference but is closer to an enthalpy difference, i.e. the entropic part is neglected. Considering the reaction under study, however, we can expect the role of entropy to be minor and the electronic energy difference to be a reasonable estimate of the free energy difference.
The reaction can then be represented by:
[A] --> [TS] --> [B]
5.6 kcal/mol,the difference between [TS] and [A], was obtained. The difference in energy [B]-[A] is actually the 1.8 kcal/mol mentionned in my previous e-mail, and it indicates that [B] is actually lower in energy. This leads to a ratio [A]/[B] of about 0.05, if I am not mistaken. So [A] and [B] are more or less isoenergetic.
Moreover, this ratio does not tell anything about the kinetics of the reaction. As We know, although the ratio does not change with time, A and B interconvert permanently with each other in a dynamic equilibrium. A barrier of 5.6 kcal/mol leads to a kinetic constant of about 10^(8), about several millions of interconversion per second for each molecule.
Hence, the answer for the question is: it can flip.
After figureing out the problem, I reprepared the ligand with alternative configruation and finally the RMSD for the ligand pose is quite close to the one in crystal structure, withRMSD<1.5Å.
https://blog.sciencenet.cn/blog-355217-693715.html
上一篇:Membrane permeability
下一篇:CHARMM TIP3P model or classic TIP3 model?