博文
J EXP BOT:基于基因的GWAS揭示大麦开花时间调控的相关基因
||
Gene-set association and epistatic analyses reveal complex gene interaction networks affecting flowering time in a worldwide barley collection
First author: Tianhua He; Affiliations: Murdoch University (默多克大学): Murdoch, Australia
Corresponding author: Chengdao Li
Single-marker genome-wide association studies (GWAS) have successfully detected associations between single nucleotide polymorphisms (SNPs) and agronomic traits such as flowering time and grain yield in barley. However, the analysis of individual SNPs can only account for a small proportion of genetic variation, and can only provide limited knowledge on gene network interactions. Gene-based GWAS approaches provide enormous opportunity both to combine genetic information and to examine interactions among genetic variants. Here, we revisited a previously published phenotypic and genotypic data set of 895 barley varieties grown in two years at four different field locations in Australia. We employed statistical models to examine gene–phenotype associations, as well as two-way epistasis analyses to increase the capability to find novel genes that have significant roles in controlling flowering time in barley. Genetic associations were tested between flowering time and corresponding genotypes of 174 putative flowering time-related genes. Gene–phenotype association analysis detected 113 genes associated with flowering time in barley, demonstrating the unprecedented power of gene-based analysis. Subsequent two-way epistasis analysis revealed 19 pairs of gene×gene interactions involved in controlling flowering time. Our study demonstrates that gene-based association approaches can provide higher capacity for future crop improvement to increase crop performance and adaptation to different environments.
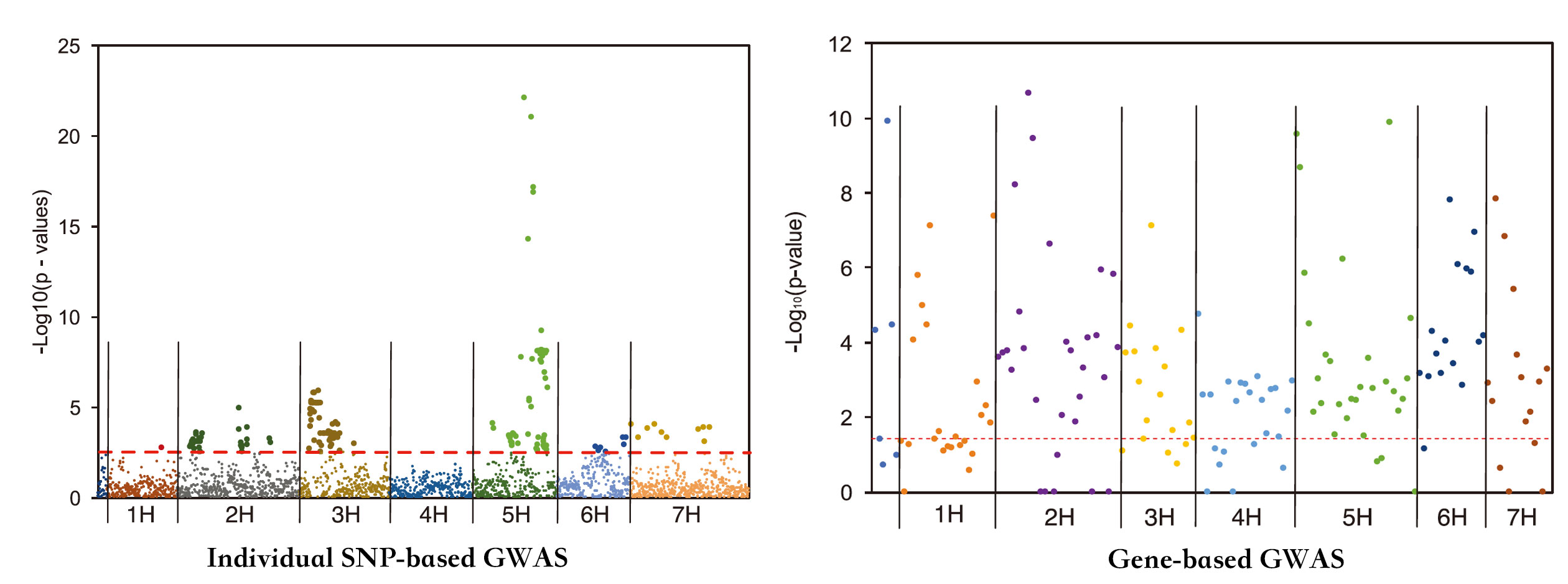
在大麦中,单标记的全基因组关联分析GWAS已经成功检测到了单核苷酸多态性SNP与开花时间、籽粒产量等农艺性状之间的相关性。然而,仅用SNP进行分析只能解释很小一部分的遗传变异,并且不能对基因网络互作进行深入研究。而基于基因的GWAS方法为整合遗传信息和遗传变异之间的互作研究提供了巨大的契机。本文中,作者重新分析了之前报道的895份大麦资源的表型和遗传数据,这些2年生大麦种植在澳大利亚四个不同的实验地点。作者采用统计模型研究基因与表型之间的关联,同时通过双向上位分析增加发现新基因的能力,这些新基因定义为对于大麦开花时间具有显著影响的基因。基因与表型的互作分析鉴定了113个基因与大麦的开花时间相关,说明基于基因的GWAS分析是十分有效的。后续的双向上位分析揭示了19对基因与基因的互作参与了对于大麦开花时间的调控。本文的研究揭示了基于基因的关联分析可以用在将来的作物育种项目中,进一步增加作物对于不同环境的表现与适应。
通讯:Chengdao Li(http://profiles.murdoch.edu.au/myprofile/chengdao-li/)
个人简介:浙江大学,学士;澳大利亚阿德莱德大学,博士;加拿大萨斯喀彻温大学,博士后。
研究方向:大麦的遗传育种改良。
doi: https://doi.org/10.1093/jxb/erz332
Journal: Journal of Experimental Botany
Published date: August 27, 2019

https://blog.sciencenet.cn/blog-3158122-1195754.html
上一篇:J EXP BOT:水稻ABC转运体G亚组蛋白ABCG18作用于根来源细胞分裂素的长距离运输
下一篇:Nature Plants:组织特异性积累的脂质磷脂酸作用于植物的盐胁迫抗性
全部作者的其他最新博文
- • Plant Physiology:CsMADS3促进柑果中的叶绿素降解和类胡萝卜素合成(华中农业大学)
- • Molecular Plant:LBD11-ROS反馈调节作用于拟南芥的维管形成层增殖和次生生长(浦项科技大学)
- • Science Advances:根结线虫通过调控植物的CLE3-CLV1模块,促进侵染进程(日本熊本大学)
- • Nature Communications:油菜素内酯参与植物营养生长期转变的分子机制解析(浙江农林大学)
- • Current Biology:光合作用产生的蔗糖驱动侧根“生物钟”(德国弗莱堡大学)
- • PNAS:花同源异型基因在叶中被抑制、花中被激活的分子机制(南卡罗来纳大学)