博文
SSB | 新加坡科技研究局A*STAR:经济高效的混合长短读长测序组装法描绘高GC含量的替代链霉菌宿主用于天然产物开发
|
近日,新加坡科技研究局A*STAR介绍了一种经济高效的混合长短读长测序组装法,用于高GC含量的链霉菌基因组测序,为加速天然产物的发现,提供了一种可行的测序组装工作流程。相关工作以“Cost-effective hybrid long-short read assembly delineates alternative GC-rich Streptomyces hosts for natural product discovery”为题发表在Synthetic and Systems Biotechnology期刊上。
研究亮点:
A cost-effective genome sequencing approach for GC-rich Streptomyces is presented.
Hybrid assembly improves BGC annotation and identification.
A new species, Streptomyces sydneybrenneri, identified by taxonomic analysis.
Genomes of 8 Streptomyces species are reported and analysed in this study.
With the advent of rapid automated in silico identification of biosynthetic gene clusters (BGCs), genomics presents vast opportunities to accelerate natural product (NP) discovery. However, prolific NP producers, Streptomyces, are exceptionally GC-rich (>80%) and highly repetitive within BGCs. These pose challenges in sequencing and high-quality genome assembly which are currently circumvented via intensive sequencing. Here, we outline a more cost-effective workflow using multiplex Illumina and Oxford Nanopore sequencing with hybrid long-short read assembly algorithms to generate high quality genomes. Our protocol involves subjecting long read-derived assemblies to up to 4 rounds of polishing with short reads to yield accurate BGC predictions. We successfully sequenced and assembled 8 GC-rich Streptomyces genomes whose lengths range from 7.1 to 12.1 Mb with a median N50 of 8.2 Mb. Taxonomic analysis revealed previous misrepresentation among these strains and allowed us to propose a potentially new species, Streptomyces sydneybrenneri. Further comprehensive characterization of their biosynthetic, pan-genomic and antibiotic resistance features especially for molecules derived from type I polyketide synthase (PKS) BGCs reflected their potential as alternative NP hosts. Thus, the genome assemblies and insights presented here are envisioned to serve as gateway for the scientific community to expand their avenues in NP discovery.
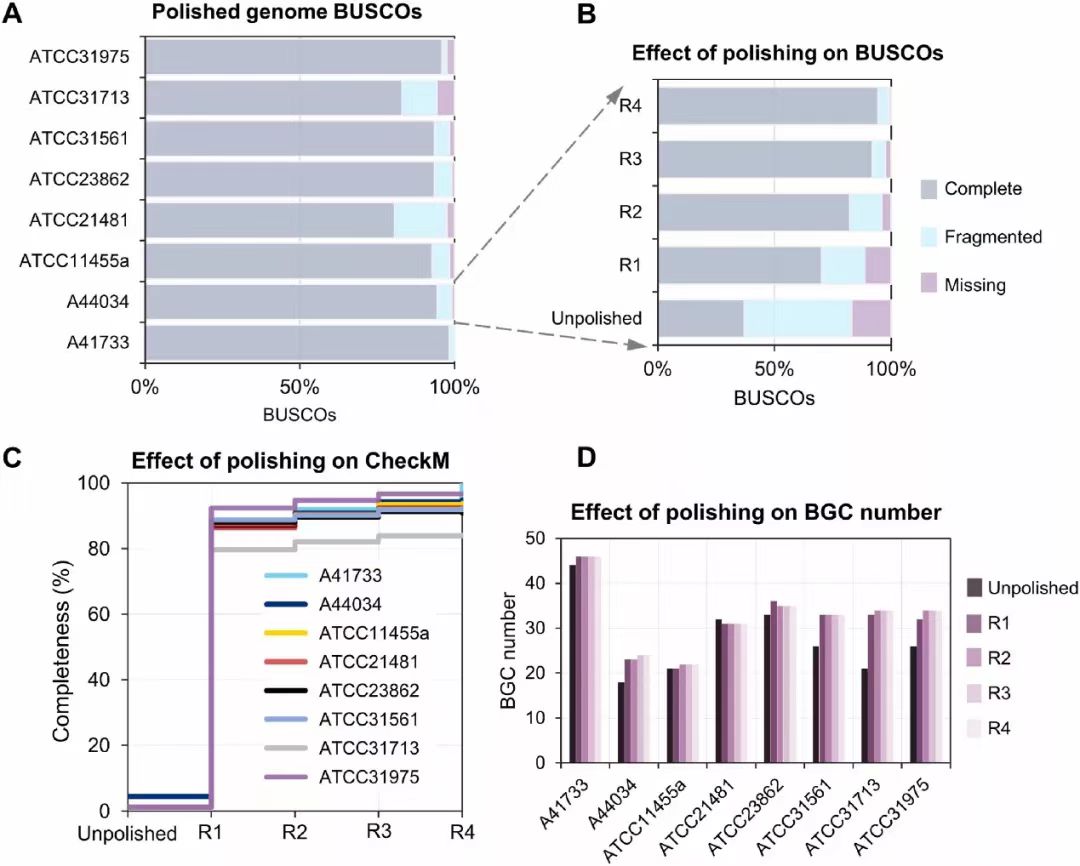
Fig. 1. Effect of polishing long read assemblies on genome quality. (A) Percentage lineage-specific completeness and contamination levels in polished assemblies as assessed by BUSCO. (B) Effect of polishing on completeness and contamination. (C) Effect of polishing on completeness in polished assemblies as assessed by CheckM. (D) Effect of polishing on number of BGC identified in assemblies. R1, R2, R3 and R4 in (B–D) correspond to the number of rounds of polishing the long read assemblies with short reads using Pilon (v1.24).
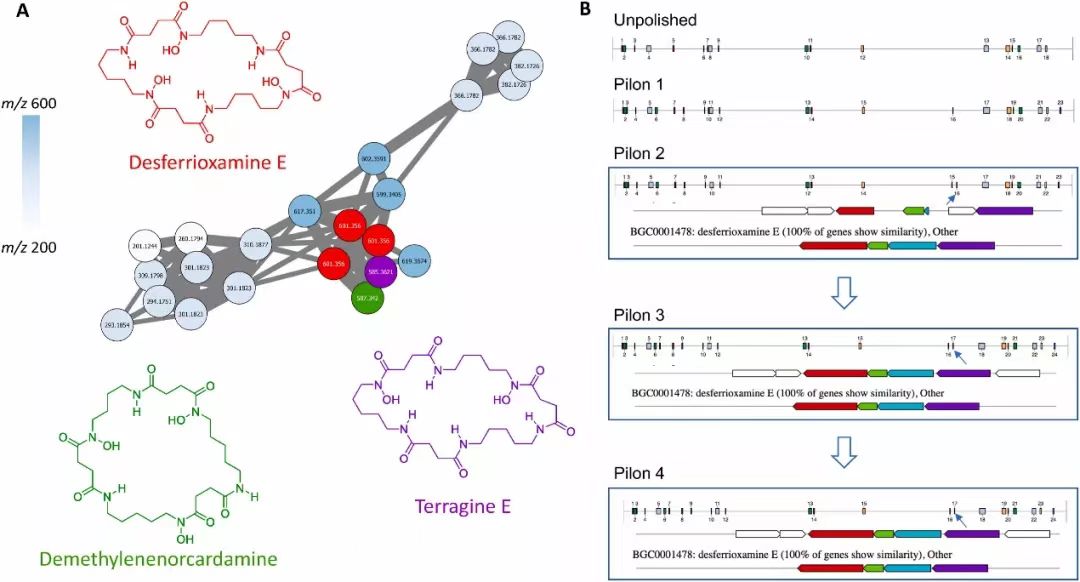
Fig. 2. Molecular networking cluster of Desferrioxamine E, and its putative biosynthetic gene cluster (BGC) in Streptomyces sp. A44034.(A) Desferrioxamine E and its analogs as determined by their MS/MS spectra and molecular networking (B) Revolution of annotated BGCs with increasing polishing rounds. Overview of clusters and their relative positions on the genome, along with multi-gene alignment as analysed using antiSMASH (v6.0) are shown. Blue arrows are used to annotate location of desferrioxamine BGC. Gene cluster table of the putative BGC is available in Table S5 of the paper. Pilon 1–4 refers to the assembly after 1–4 rounds of polishing, respectively.
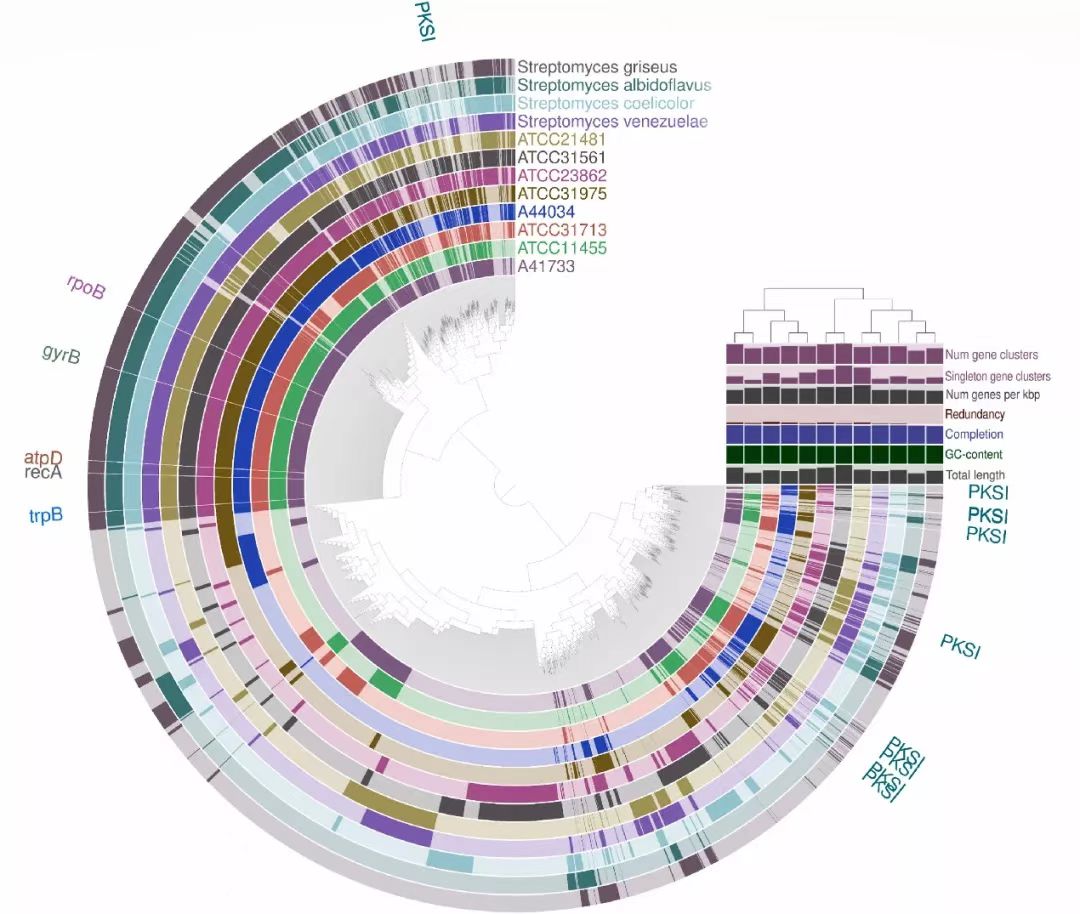
Fig. 3. Pan-genomic analysis of the Streptomyces strains. The circular strip represents the presence/absence of gene clusters in each of the strains. PKSI: type I PKS; rpoB, gyrB, atpD, recA and trpB are housekeeping genes of Streptomyces. Bar plots at the end of each strip represents additional layers summarizing pan-genomic statistics of strains. ‘Num gene clusters’ represents the total number of gene clusters in strains, ‘Singleton gene clusters’ are orphan clusters which do not have homologs in other strains. Dendrogram is constructed using gene cluster presence/absence and Ward clustering. Tree over the layers is constructed based on Euclidean distance between the corresponding values for the strains using hierarchical clustering. Numbers corresponding to different layers are provided in detail in Table S6.
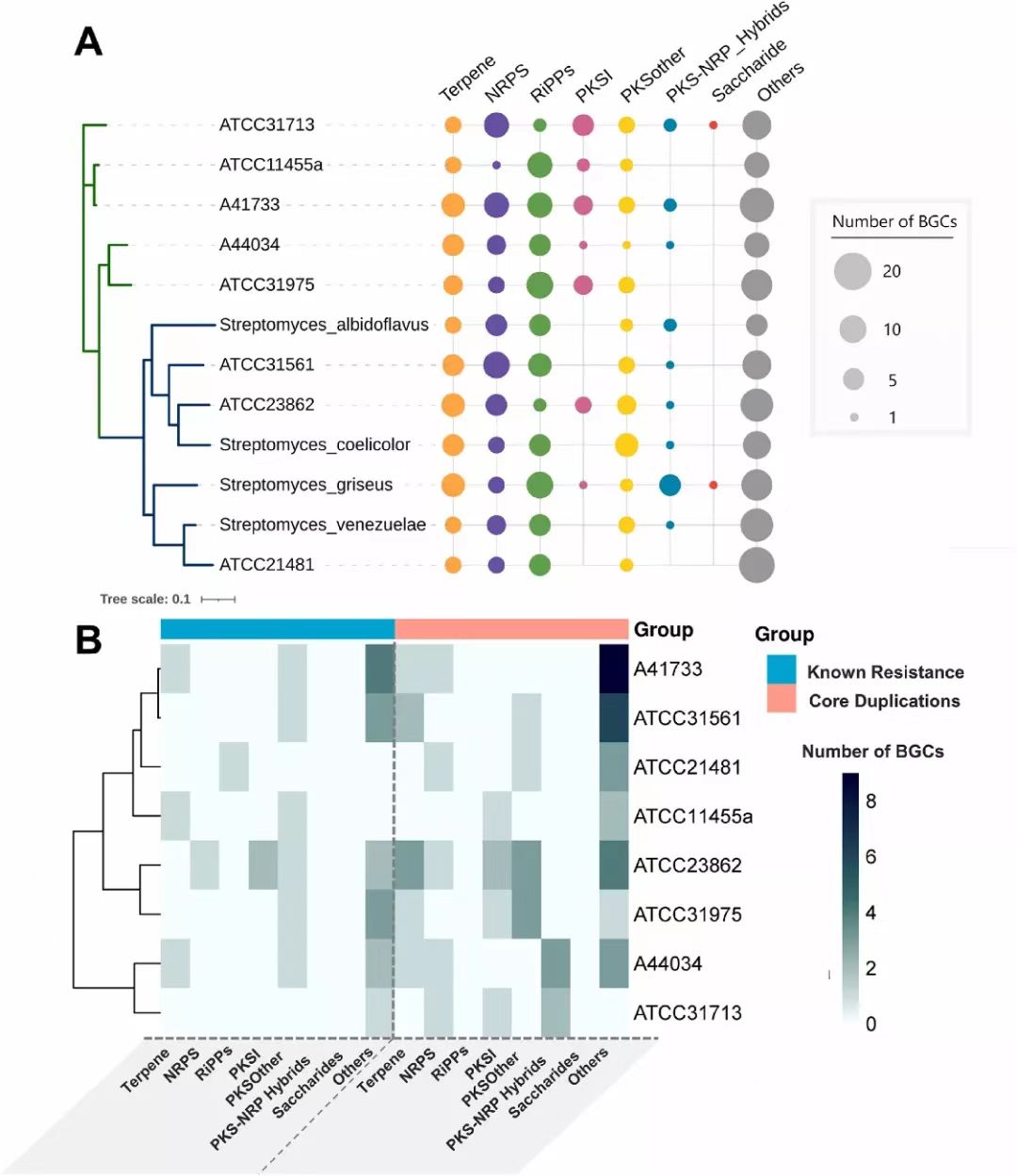
Fig. 4. Mining BGCs and resistance mechanisms (A) Enrichment of BGC classes in different Streptomyces strains. The size of the circle is proportional to number of BGCs harboured by the corresponding strain. The tree is constructed based on whole-genome phylogeny (bootstrap = 100). The clade overrepresenting model Streptomyces is coloured blue, while the strains in this study mostly fall under the green clade. More details are provided in Table S7. (B) Presence of known-resistance mechanisms and duplicated core genes inside BGCs of different classes as identified by ARTS analysis (v2.0). NRPS: Non ribosomal peptide synthase, RiPPs: Ribosomally synthesized and post-translationally modified peptides, PKSI: Type I PKS, PKSother: PKS other than type I PKS, PKS-NRP_hybrids: Hybrid PKS-NRPS, Others: Cluster that does not fit into any of categories shown.
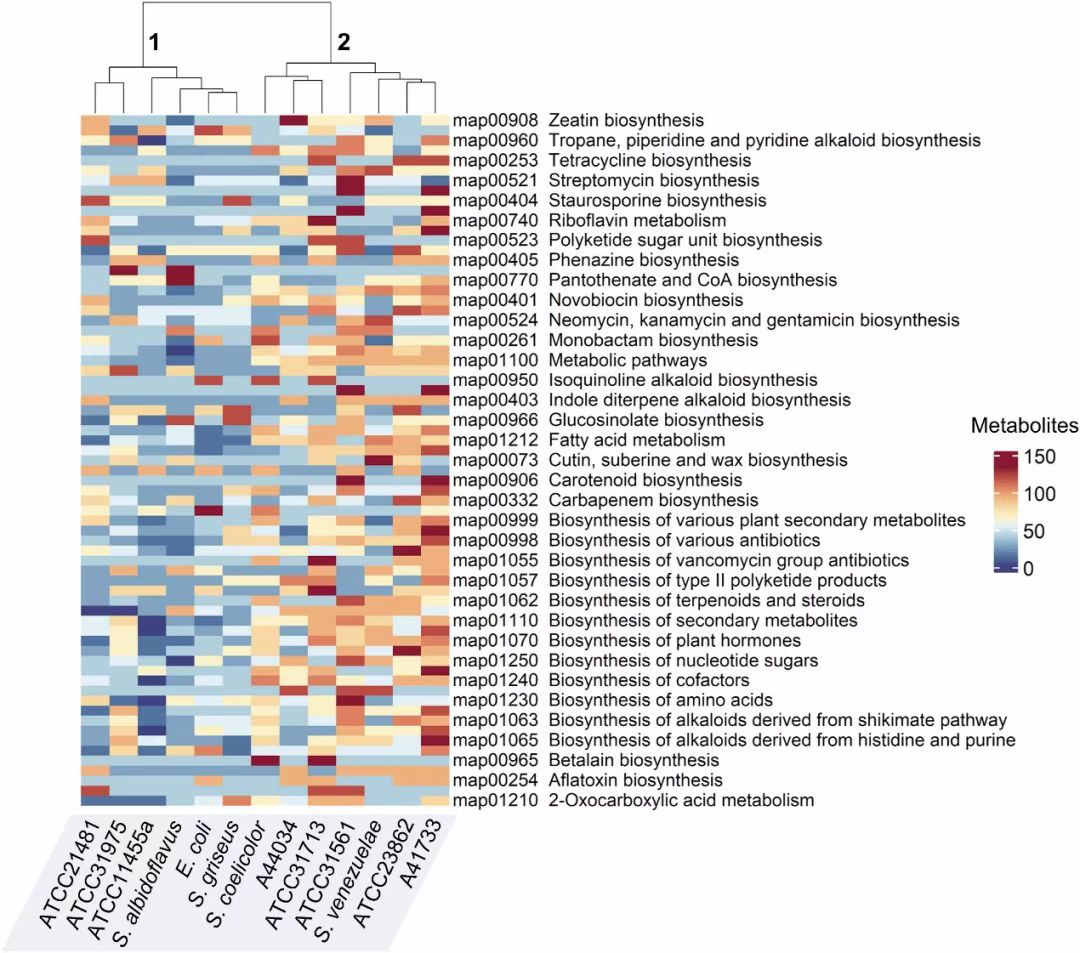
Fig. 5. Biochemical network-guided metabolic pathway enrichment. The heatmap showcases enrichment of different KEGG pathway maps related to NP biosynthesis in the 8 strains, 4 prototypical Streptomyces and the widely used heterologous host, E. coli. The color scale corresponds to number of metabolites in each KEGG pathway, red being ‘high’ and blue corresponding to ‘low’ values. Labels ‘1’ and ‘2’ at the top represent two distinct clades showing significant differential enrichment in NP biosynthetic pathways (p-value = 1.2055e-04, chi-squared = 14.78, Kruskal-Wallis test). The ‘map’ IDs denote KEGG pathways. The dendrogram was generated using hierarchical clustering. Detailed numbers can be found in Table S16.
Cost-effective hybrid long-short read assembly delineates alternative GC-rich Streptomyces hosts for natural product discovery
Elena Heng, Lee Ling Tan, Dillon W.P. Tay, Yee Hwee Lim, Lay-Kien Yang, Deborah C.S. Seow, Chung Yan Leong, Veronica Ng, Siew Bee Ng, Yoganathan Kanagasundaram, Fong Tian Wong, Lokanand Koduru.
https://doi.org/10.1016/j.synbio.2023.03.001
Synthetic and Systems Biotechnology是高质量国际开放获取期刊,创刊于2016年。期刊覆盖合成生物学、系统生物学以及生物医药等领域。期刊现已被SCIE、EMBASE、PubMed Central、Scopus、CSCD等重要数据库收录。
2021 Impact Factor: 4.692; 5-Year Impact Factor: 5.23; JCR分区Q2
2021 CiteScore: 6.60, 位列学科Q1区
2022中科院分区生物学大类Q2区;生物工程与应用微生物小类Q1区
入选2019年中国科技期刊卓越行动计划高起点新刊项目
https://blog.sciencenet.cn/blog-3496796-1383670.html
上一篇:线下会议 | 第二届先进生物制造国际会议(ICAB 2023)
下一篇:BAM|中山大学附属口腔医院龚启梅和韦曦团队:低氧预处理牙髓干细胞来源小细胞外囊泡有效缓解炎症性骨溶解进程